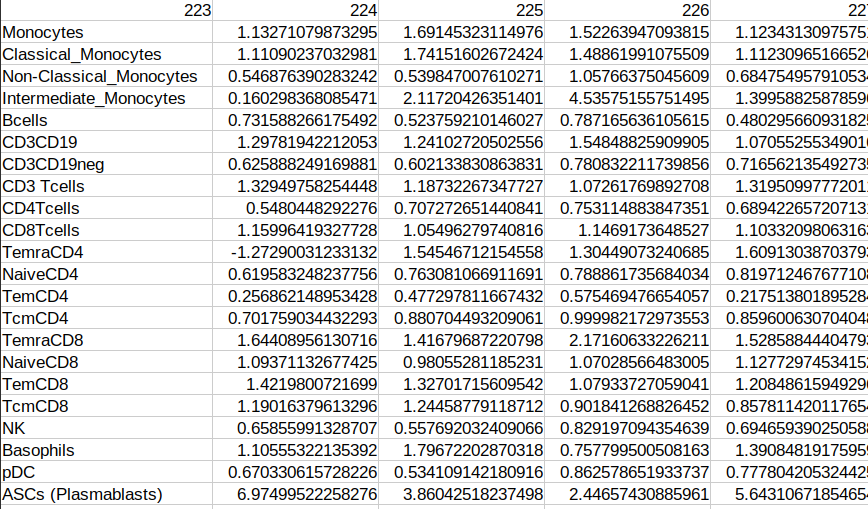
Are the headers of the columns in file pbmc_cell_frequency_batchCorrected_data (and probably others) misaligned? The 223 should not be on top of Monocytes, but on top of 1.1327. To me, the headers appear all left shifted (which causes the last column to be without a header). I’ve attached an image.
The processed data files are saved in matrix formats where the first column contains the row names (feature names). This is why you may notice that columns appear shifted to the left. When reading the processed files, both training and test files, it’s important to consider the first column as the row names.